Forum Replies Created
-
AuthorPosts
-
sychoi
ParticipantHi Dan,
you can try cutting through the surfaces by pressing keys “x” “y” or “z” on the surface viewer.
x : sagittal
y : coronal
z : axial
The surface will cut through where ever your crosshairs are located on the volume slices.sychoi
ParticipantHi Dan, our atlases will be found with your installation files. for example
for windows:
C:\Program Files\BrainSuite18a\svregfor mac
/Applications/BrainSuite18a/svregyou will see a directory for the BCI-DNI_brain atlas and BrainSuiteAtlas1. I recommend the BCI-DNI_brain atlas.
drag and drop files ending in:
.bfc.nii.gz (or BCI-DNI_brain.nii.gz for inclusion of skull and scalp)
.left.mid.cortex.dfs
.right.mid.cortex.dfs
.all.dfc (right and left will only load one at a time)
and under File–>open–>label volume
select the “BCI-DNI_brain.label.nii.gz” fileTo give you a few tips:
you can learn about navigation controls here: http://brainsuite.org/interface/
you can click anywhere on the volume slices or double click anywhere on the surface and the name of the region will appear on the bottom of the interface. There’s also some fun things you can do for visualization by opening up the mask toolbox.
You can see the names and description of the sulcal curves (.dfc files) by opening up the sulcal toolboxclick around, experiment and have fun. Let us know if you have any other questions.
sychoi
ParticipantI think what you’ve tried so far is great.
It seems for a closed surface, rendering a surface using the svreg labels are your best bet so far.
Can you specify what was inaccurate about your surfaces?Note also that your svreg labels will be subdelineated by GM and WM labels for each individual cortical label. ROI ID labels for GM starts with “1” and for WM “2”
example: 1120: R. Superior frontal gyrus (WM) 2120: L. Superior frontal gyrus (WM)I’m not sure how we can help you more with the given information. What are you trying to calculate exactly?
sychoi
ParticipantSorry I didn’t completely understand your question but are you asking how to load your .nii file?
You can simply open brainsuite and drag and drop the file directly onto the gui.
Or you can go to File–> Open volume.sychoi
Participantthe subject.pvc.label.nii.gz will have GM/WM/CSF labels only.
Then the subject.skull.label.nii.gz will have skull, scalp, space and brain labeled.detailed information can be found here: http://brainsuite.org/processing/surfaceextraction/skull-and-scalp/
July 7, 2017 at 3:03 pm in reply to: Merging external ROI files (nii format) to Label description file #762sychoi
ParticipantThere is a number of different ways to do this interactively on the GUI or automated using matlab.
Are you trying to add ROIs already created in subject or atlas space?
or are you trying to add new ROIs by manually drawing them in the GUI?sychoi
ParticipantIt looks like the bvec file is not in the format that can be read by BDP
Here is a sample of what BDP looks for: http://brainsuite.org/examples/siemens_20.txtYou might have to reformat it to this or I would suggest using dcm2nii to convert your dicoms to nifti
sychoi
ParticipantThe hemi.label file is an intermediate file that we use to roughly identify regions of the brain. If you load the hemi.label.nii.gz file in brainsuite, you will see that it labels the right and left brainstem, cerebrum, and a general subcortical area.
You can take a look at your tissue classification results by opening the pvc.label.nii.gz or pvc.frac.nii.gz
Here’s a video where I explain the brainsuite outputs: http://brainsuite.org/video-tutorials/checking-outputs/In order to get calculations of GM/WM/CSF and total volume, run your subject through SVReg which can be run on the GUI or command line. (instructions found here: http://brainsuite.org/processing/svreg/)
it will output a roi.stats.txt file which outputs volume measurements which you can open in excel. the 1st line will give you total CSF volume, 2nd: GM Vol and 3rd WM Vol.
It will also list regional measurements and you can find the description of the ROI IDs by opening the brainsuite_labeldescription.xmlIt looks like for this subject I get
CSF: 272.569
GM: 682.809
WM:521.965
GM+WM: 1204.774BrainSuite uses partial volume tissue classification which considers that some voxels are composed of mixed tissue and assigns it that way. So a single voxel can be calculated as 40% GM and 60% WM which is why you get voxels labeled as GM/WM or GM/CSF. This can give you more accurate measurements.
sychoi
ParticipantHi BrainSuite does not support image reorientation.
If ImageJ did not work for you, other software packages such as Rview and FSL also provide this module. a quick google search will also direct you to some matlab packages.
Good luck.
sychoi
Participantyou can find the ROI ID and descriptions in the an xml file called “brainsuite_labeldescription.xml” in your atlas folder. The atlas directory can be found in your brainsuite installation directory:
Windows:
C:\Program Files\BrainSuite16a1\svreg\BCI-DNI_brain_atlas\brainsuite_labeldescription.xml
Mac:
/Applications/BrainSuite16a1/svreg/BCI-DNI_brain_atlas/brainsuite_labeldescription.xmlsychoi
ParticipantYou might not need to create a custom atlas in your case because a lot of the necessary components is already provided.
Quick question first, are any of these labels cortical? Or are all subcortical and/or outside the cerebrum?
sychoi
ParticipantBrainSuite’s surfaces are in .dfs format which you can load in matlab. Here are the instructions: http://brainsuite.org/processing/svreg/matlab/
There are additional tricks you can find at the end of this post: https://forums.brainsuite.org/forums/topic/standard-surface-rendering-file-formats/
I have never tried this myself but let me know if this works out for you.
sychoi
Participantopen the image display toolbox under View–>Image Display Properties
right click on the colobar of the volume you want to recolor and choose the color scheme of your choice. Here is a screenshot: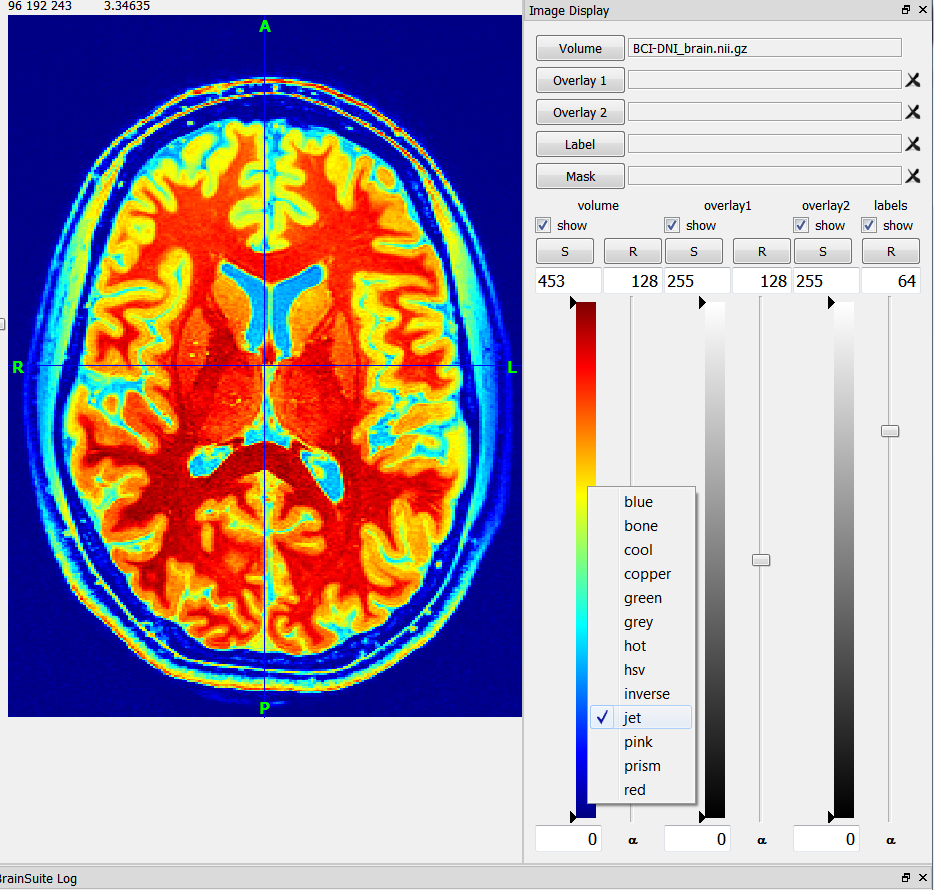
sychoi
Participantat the moment we do not support .trk file formats.
sychoi
ParticipantSorry for the late response. From dcm2nii you should be getting a nifti file, bvec and bval file for each series. If you are using a philips scanner with their stock sequences, you will have a dummy volume at the end and dcm2nii will output 6 files for each series. the 3 files mentioned and the same 3 files but starting with “x” where the dummy sequence is removed which is what you want to use.
You can find example files here: http://brainsuite.org/WebTutorialData/DWI_Feb15.zip
and explanations of file formats here: http://brainsuite.org/processing/diffusion/file-formats/ -
AuthorPosts